Gene ontology and network
- IntAct
- GeneMania: for beautiful gene networks. Indexing 2830 association networks containing 60554667 interactions mapped to 166691 genes from 9 organisms.
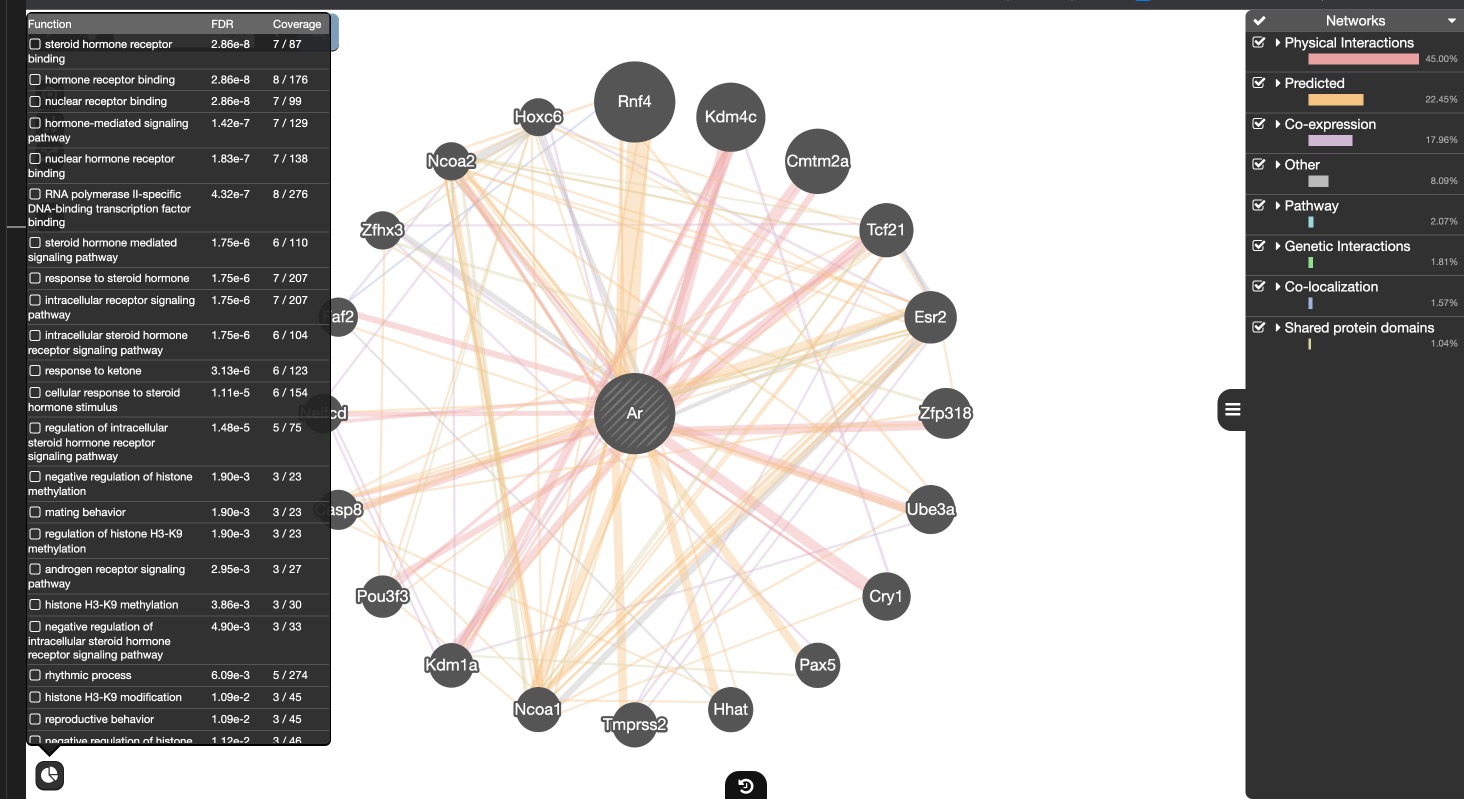
Usage case in peer reviewed paper - Pathway Commons

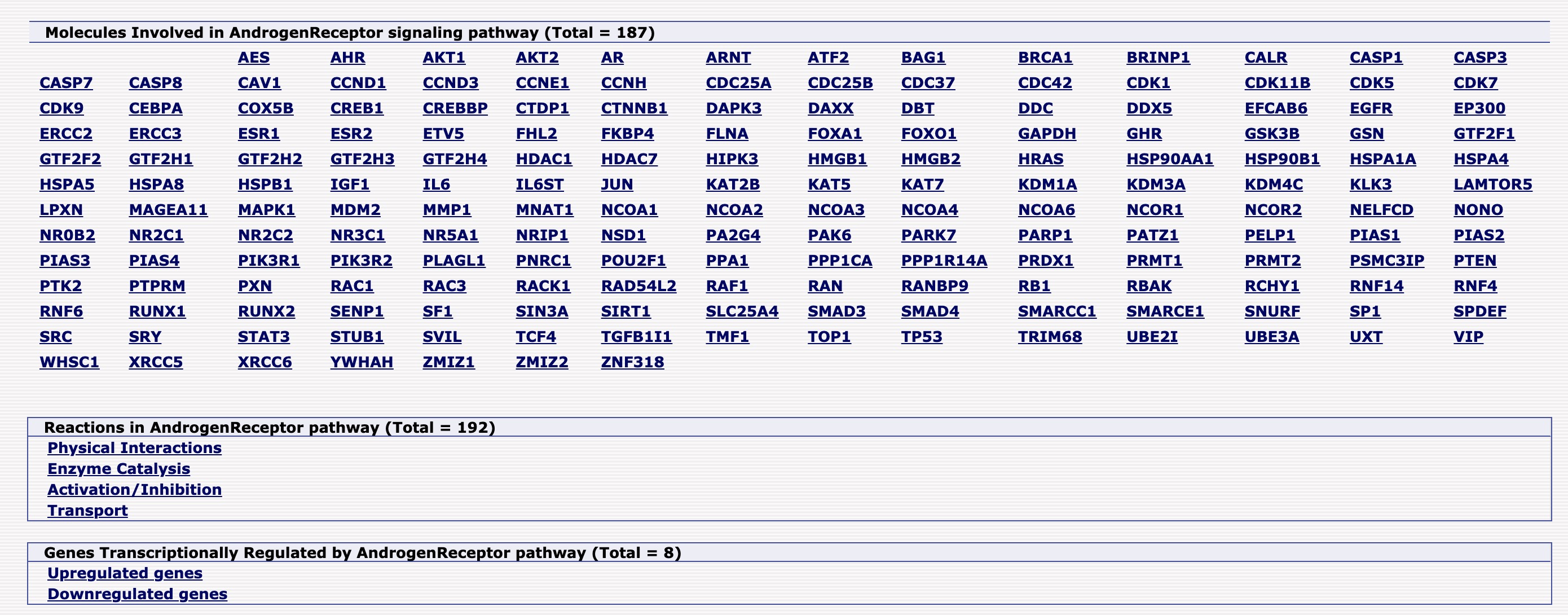
- Human transduct signaling pathways
- Mouse knowledgebase: Genes (ontology and signaling pathways), phenotypes & mutant alleles, Human-Mouse disease connection, gene expression, cre, strains & polymorphisms, homology, cancer models, …
- Enrichr Usage case in peer reviewed paper
TF regulon
- Human: Encode transcription factor targets: 22449 genes, 181 TFs and 1651393 gene transcription factor associations by binding of transcription factor near transcription start site of gene
Gene expression
- Tissue level:
- BioGPS provides an extensive annotation using gene expression from ArrayExpress, Exon array Gene Atlas, Gene expression/activity chart, Gene Expression Atlas (ArrayExpress Atlas), NCBI Gene Expression Nervous System Atlas (GENSAT), The Human Protein Atlas and a 15 human placenta studies.
- ARCHS4 provides access to gene counts from HiSeq 2000 and HiSeq 2500 platforms from GEO and Sequence Read Archive (SRA).
Cell type markers:
- Cell Ontology. CL was first developed as a platform in 2004 to collect major cell types from humans and model organisms, and has since been applied to various fields. Each CL has a permanent url.
- Panglaodb provides 13 marker genes for undefined human placental cells identified from embryonic and placental tissues without distinguishing cell types.
- CellMarker provides marker genes for 17 cell types from human placenta tissues from either single-cell sequencing, experiment, or review, are reported.
- Human protein atlas reanalyzed the first-trimester human fetal interface single-cell transcriptomic data from Vento-Tormo, et al. altogether with data from 36 other tissues. They identified 3 cell type enriched genes in cytotrophoblasts, 45 in syncytiotrophoblasts, and 26 in extravillous trophoblasts via the elevated mRNA level compared to other cell types.
- Descartes cell atlas depositing data from a peer reviewed papar.
- The Human BioMolecular Atlas Program (HuBMAP) ASCTplusB tables include information about full-term human placenta. For this database, protein and lipid biomarkers have been curated for 24 cell types based on published placental histopathologic nomenclature, and literature on cell-type specific biomarkers.
- Celltypist, especially for immune cells.
genome browser
I like WashU