all the talks are with preprints
Genome wide TF binding
SHARE-Seq
In the aim of interpreting the footprint, in my words, to build a DNA footprint database –> trans-element
<– DL (inspired by dl)
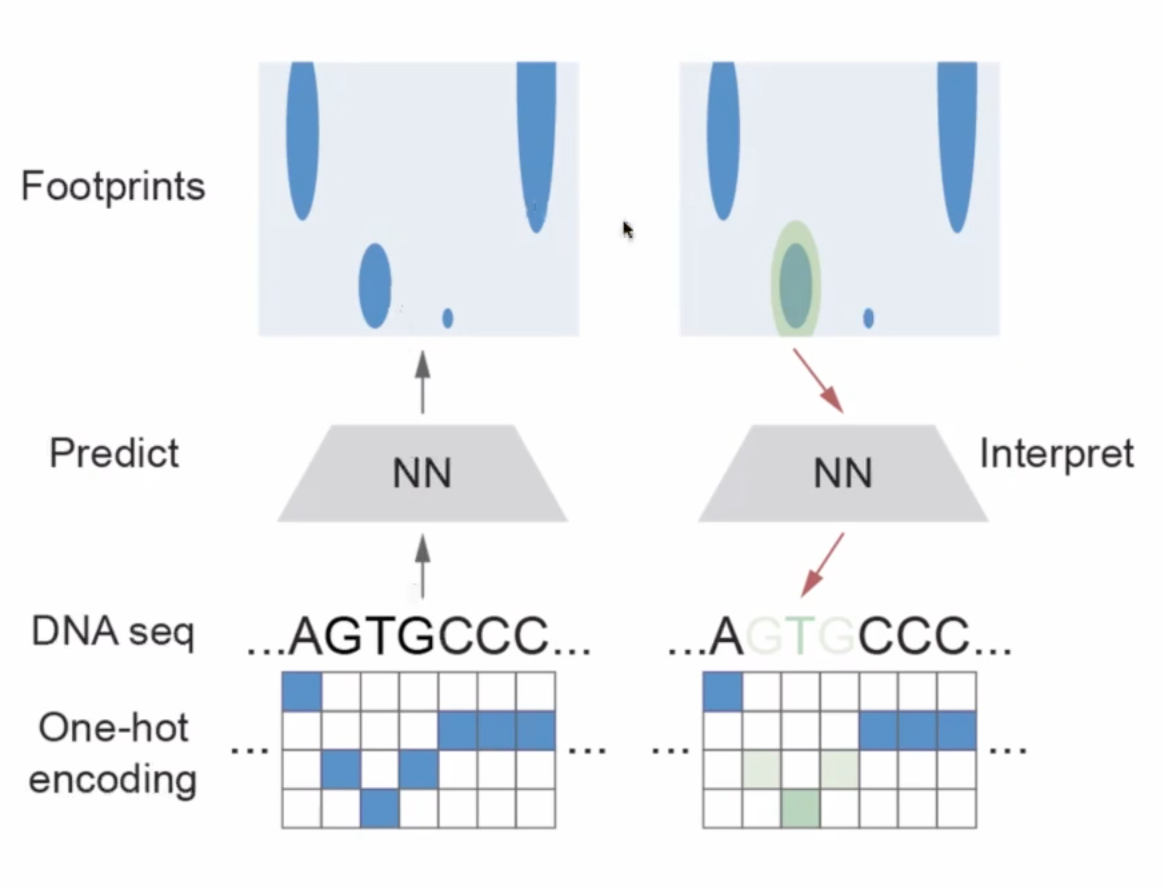
How stable are the footprint as DNA are stable?
can it output pleipotrip footprints?
the scanning params (padding)
–> use case identify TFs that drive cell aging –> nucleosomes shift
3D spatial from “raw” 2nd seq
 –> 3D reconstruction based on more raw photonic data
–> 3D reconstruction based on more raw photonic data
 –> expand the in situ seq by introducing the expanded hydrogel –> data on microns or DNA distance from lamin
–> expand the in situ seq by introducing the expanded hydrogel –> data on microns or DNA distance from lamin
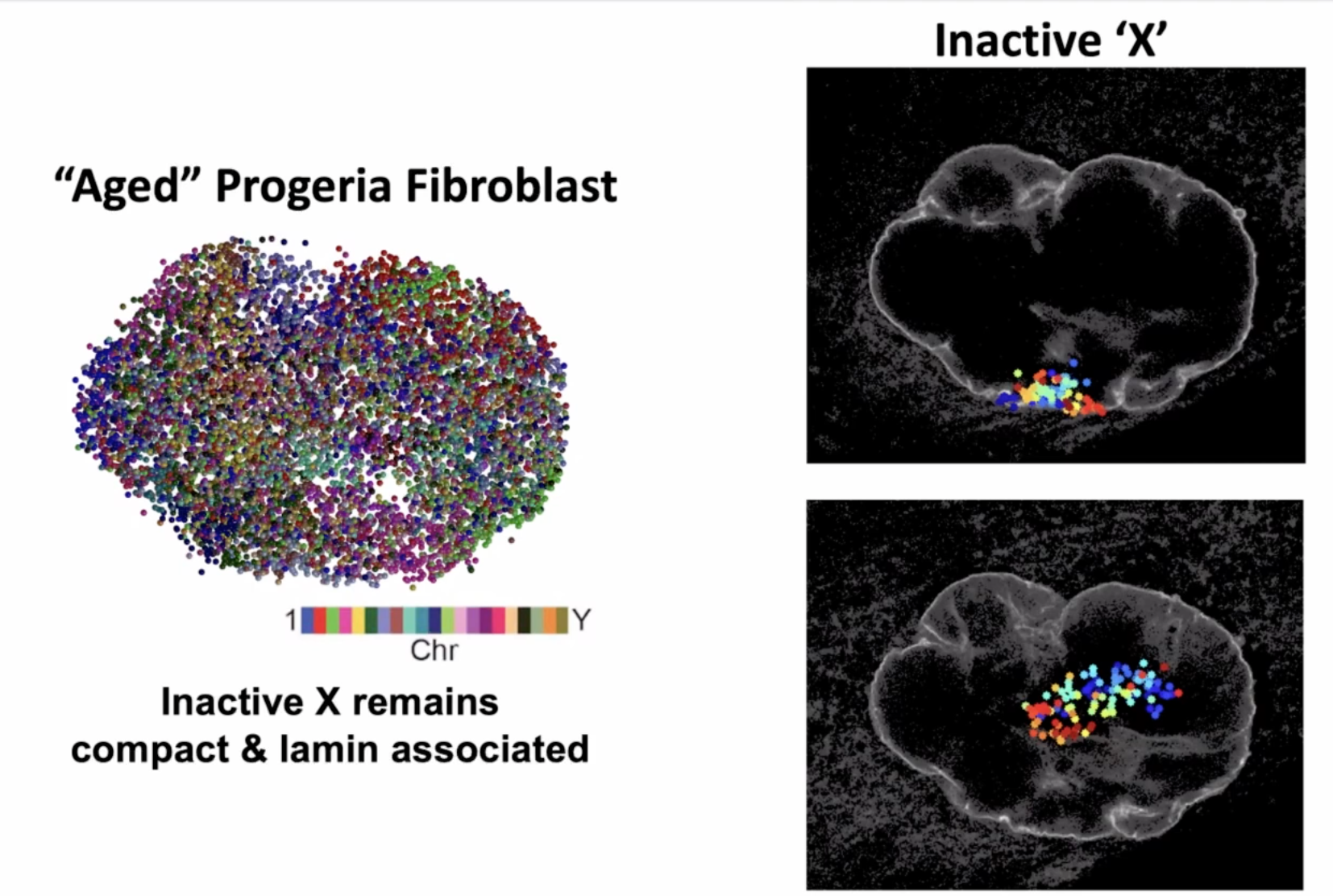
coupling with lamin to study the chromatin structure is a convention
inactive “X” as an exampled biology to be studied with this
Sumamry of chromatin resolution and techs
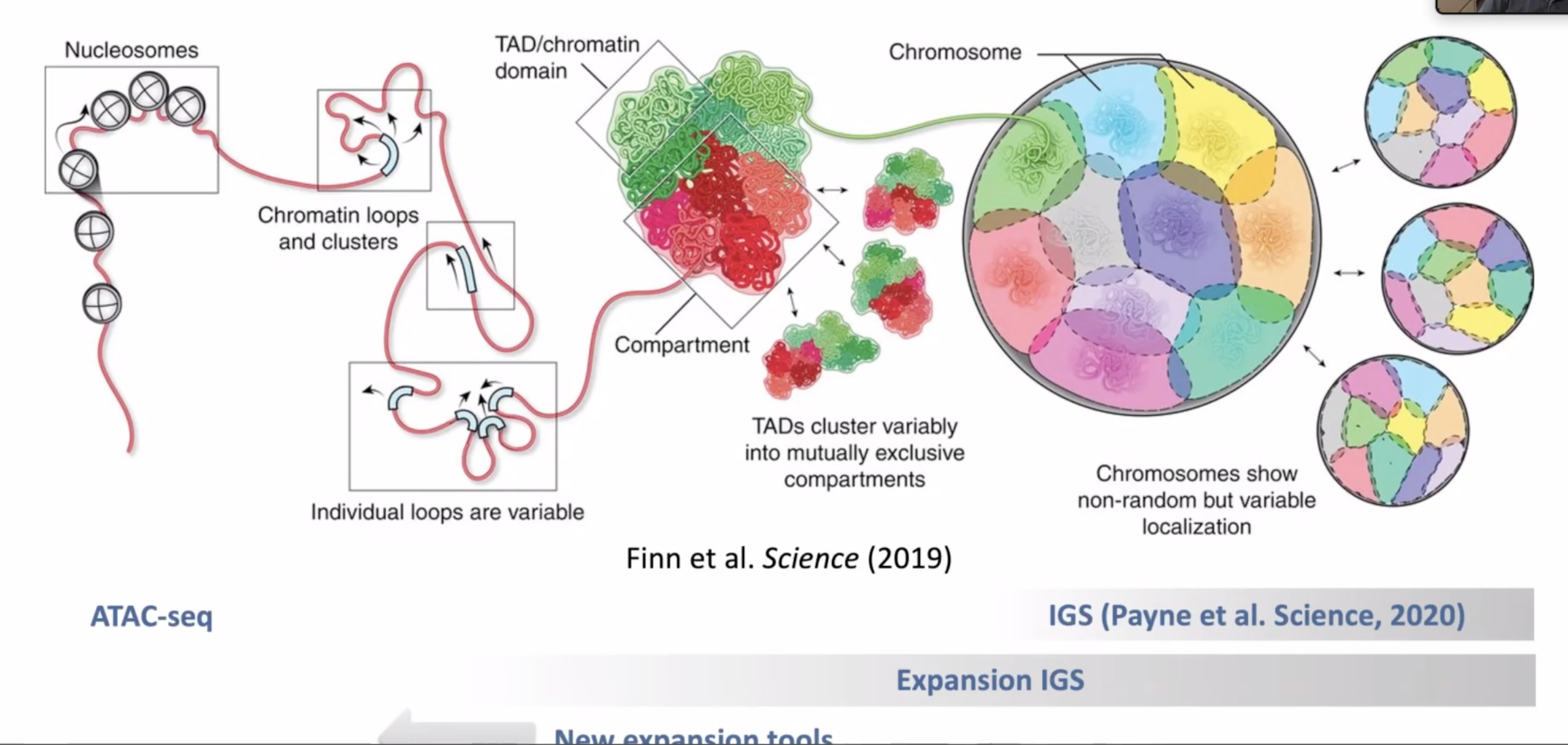
Discussion
ATAC replace ChIP
but it’s based on knowledge from ChIP
open-ST
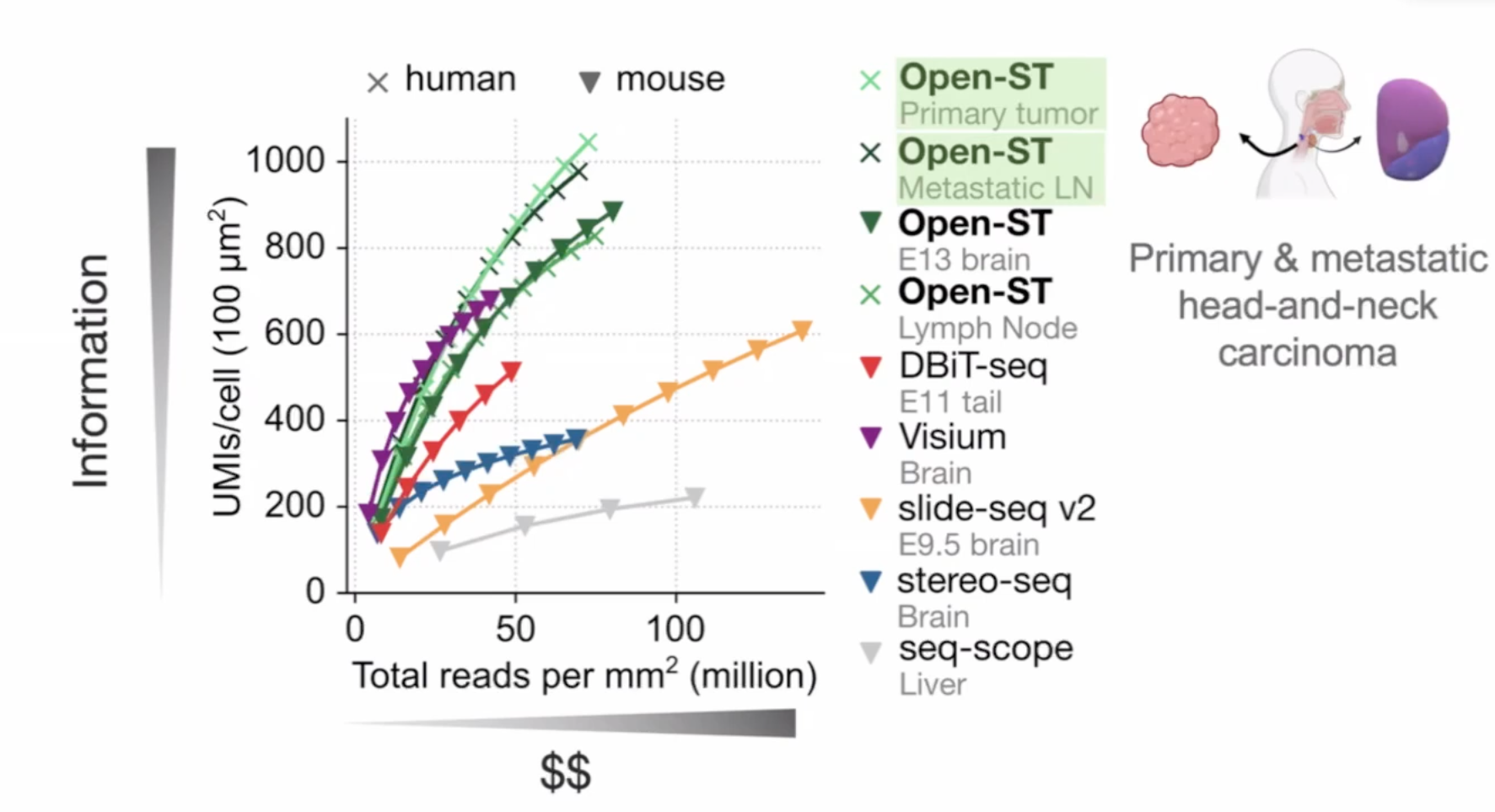
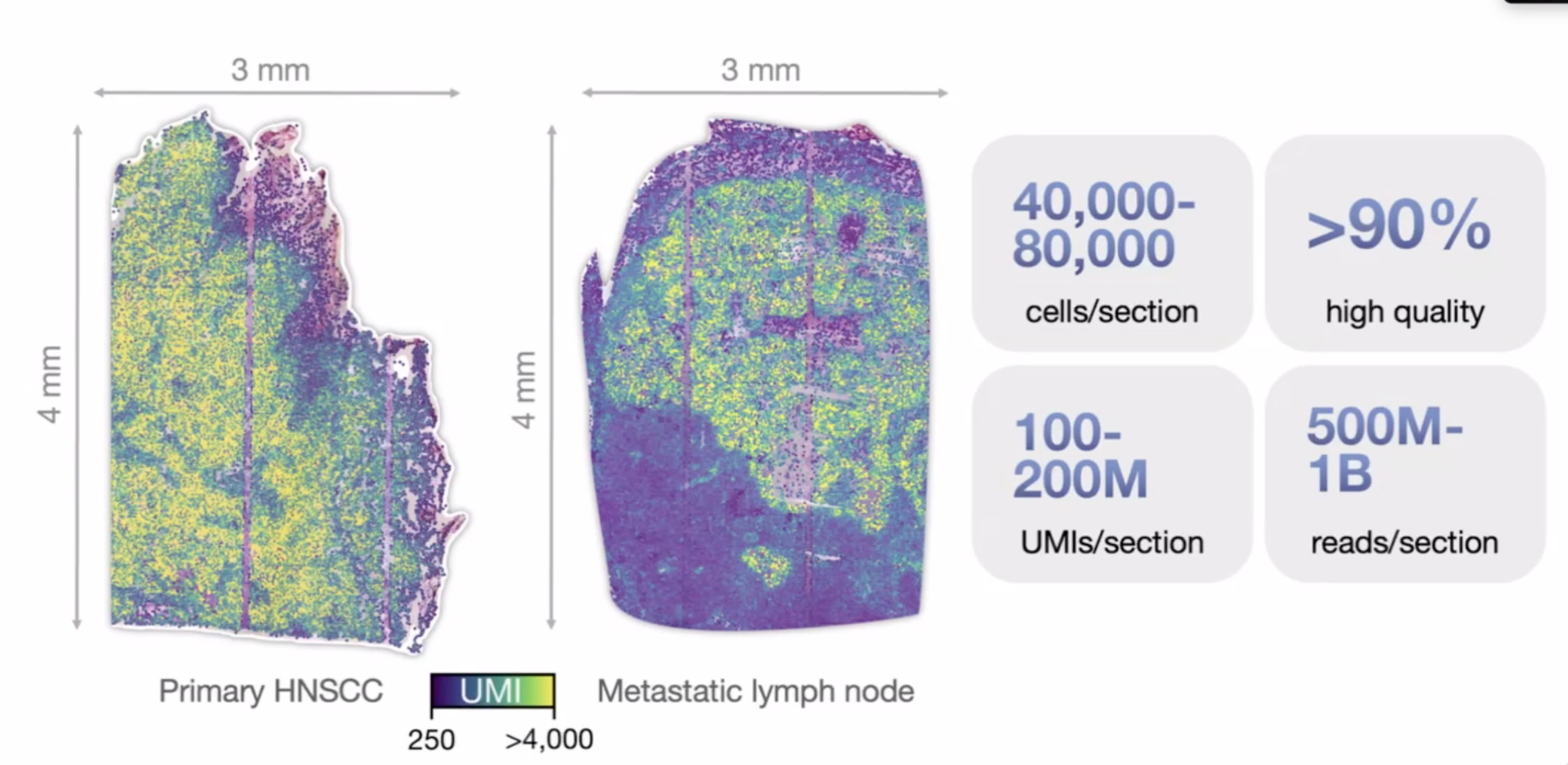
20,000 genes
from Nikolaus Rajewsky lab
“3D virtual tissue block”
module cell segment, module transcripts, map modules
biological output
3D boundary between cell types, for example, tumor cells and other cells
Discussion
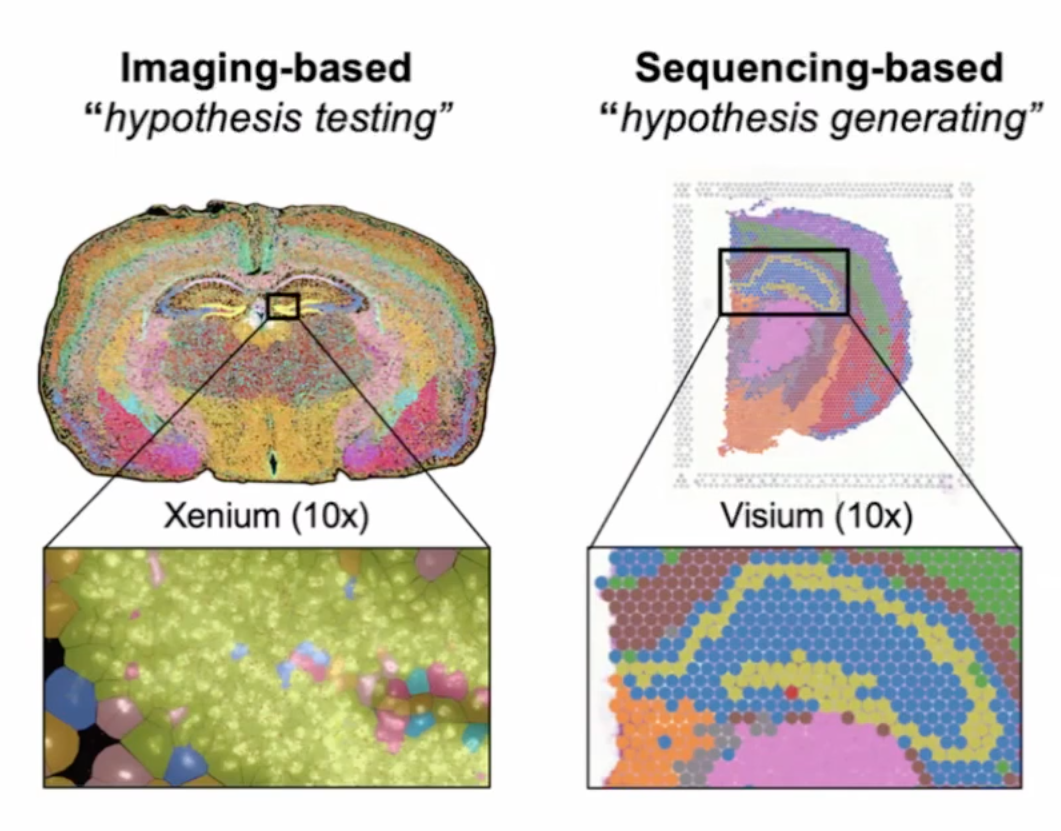
- sequencing-based has more varied complexed noises. Ideal for hypothesis generating
- imaging-based is ideal for validating the hypothesis.
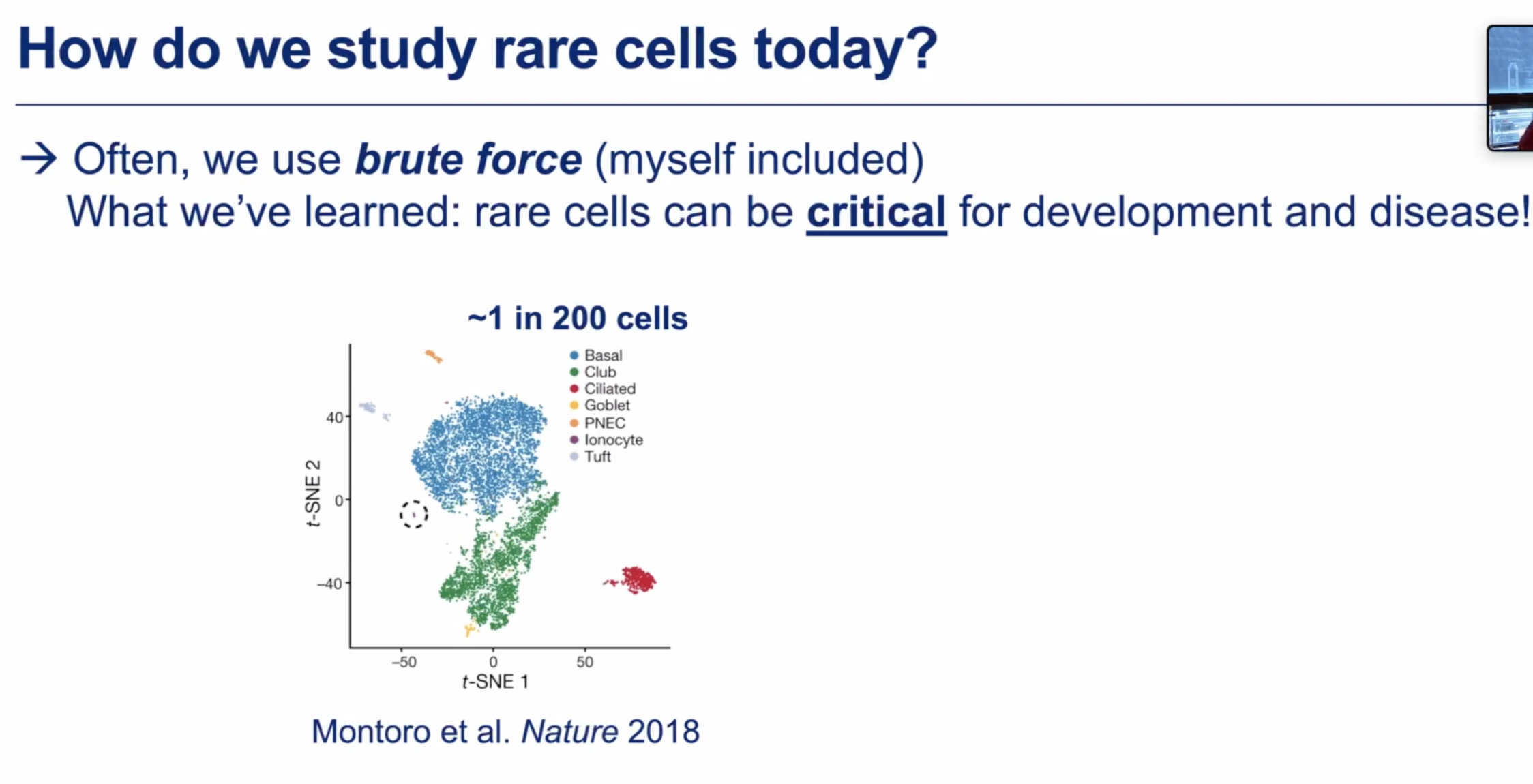
Rare cell type by na cytometry based PERFF-seq
Coleb Lareau
study strategy
how much they are differ from other non-rare type?
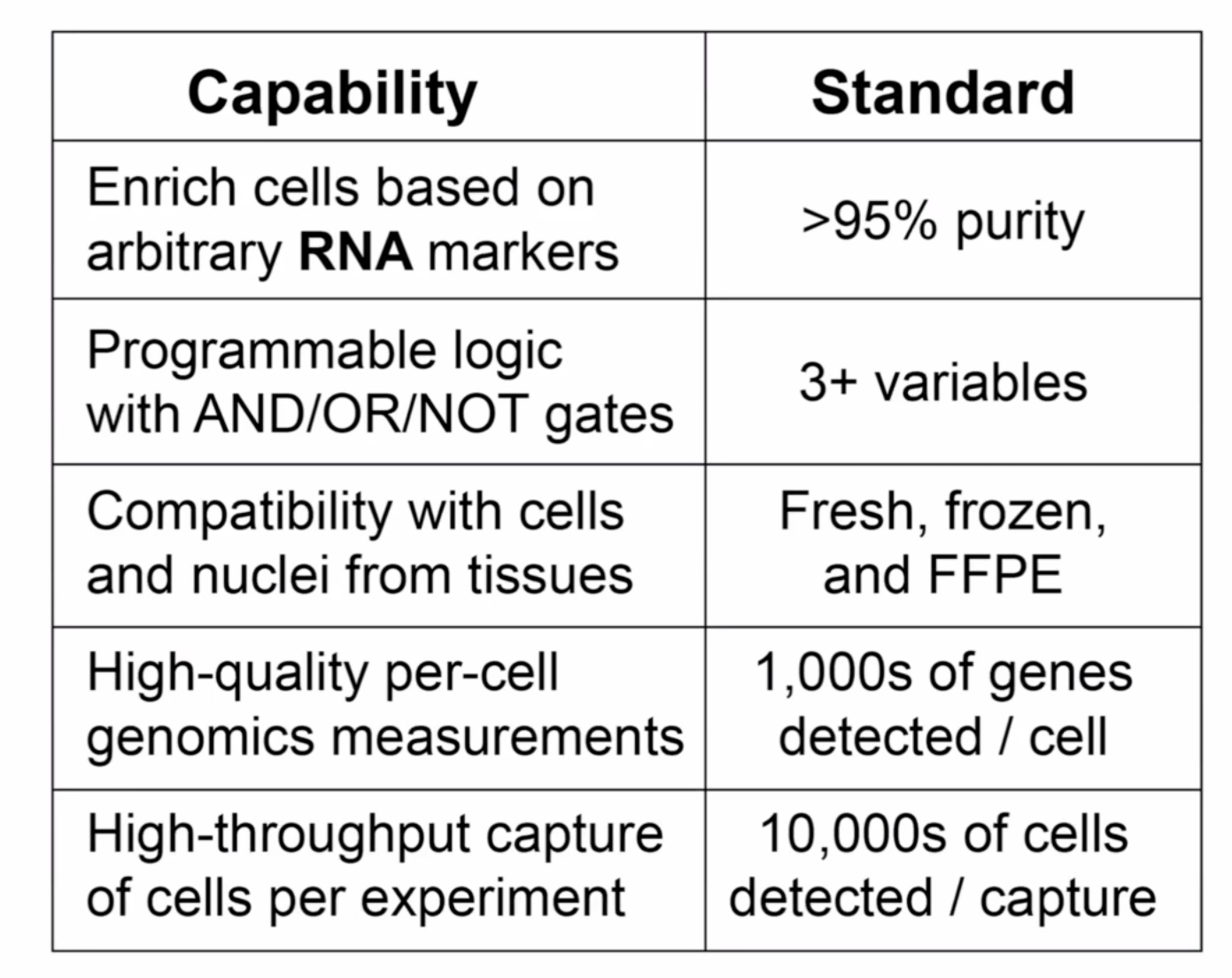
Criteria for sucessful assay
Based on
- RNA FISH
- 10x Genomics Flex scRNA-seq, where cells are fixed
I didn’t understand how seq on fixed cells bu two probes on one read
benchmark using FC/RNA cytometry enrichment
Biological output
- enrich cells with TF expression
- enrich with coding and ncRNA
- rare cell types enrichment
- FFEP and frozen tissues
Discussion
- easier with RNA vs DNA
- initiate probes? ~ size of target gene
- how many markers? limited by cell numbers but not markers
I would say is the choice of orthogonal markers
Single cell perturbation dictionaries
- Chemical (cytokine stimulation)
- Cui, wt al, Nature (2024)
I read this quite carefully already
- Cui, wt al, Nature (2024)
- Perturb-Seq
- first cytokine stimulation
- then fix and permeablize for perturb-seq (3 guides per gene for 40-60 genes)
Analyzing
- binarize perturbatoin output as in Nat Genet (2021)
- imporve to continous score
spatial and sc by Saturn and star-map
convention
- smoothing I missed how
- sc analysis neighbors are also spatial neighbors
solution
- sample from multiple neighborhoods
- smoothing
- sc analysis
spatial-resolved sc genomics of human brain by Xiaowei Zhuang
- space of cells in tissues
- space of intra-cellular
- space of chromasome
Techs
-
MERFISH > 1k genes 2019 Technology
- error-robust bc
- combinatorial label
- sequential imaging
-
MERFISH extened to 3D genomics 2020
localize DNA loci > 1k genomic loci
+ > 1k nascent RNA
+ protein
- extended to epigenomics
with antibody to Tn5 taged protein
in a gel so with proximal info
–> epigenetic markers
- thick tissue 3D merfish
200 $\mu m$
biology
- Mouse brain cell atlas. related pub: Zhang et al., PMID 38092912 (2023) somatic contact, paracrine signaling or short-range interactions compose cell-cell interaction
- Human brain cell atlas
Discussion
human mouse diff (behaviral)
- not only from dense (human < 3x mouse)
- modulize effects
- loci to genomic to get chromosome organization by imaging
- techniqually no problem to increase
- high density in nucleus as a rate limiting –> more runs, so tedious
- worth efforts
- now mega base resolution –> higher resolution, you need more runs and sts, imaging time
I didn’t think of a way to get around this density
Fixed Universal cell embeddings by Yanay Rosen
Concepts
- e.g. put cells in one universal waddington landscape
- AI e.g. from CZI
with this, we could model cellular change in silico
Represent cells with - representable expression matrix, e.g. low-dimensional embeddings - a sentence then can study with NLP - feature ordering? - a bag of RNA (no order)
I would consider this is prior-free - transformer Represent genes - Excel (redicule) - Gene2Vec - chatgpt
Solution:
Gene represent adopting an aa model ESM
Fixed:
NOT finetune, so called 0-shot
**Self-supervised
therefore
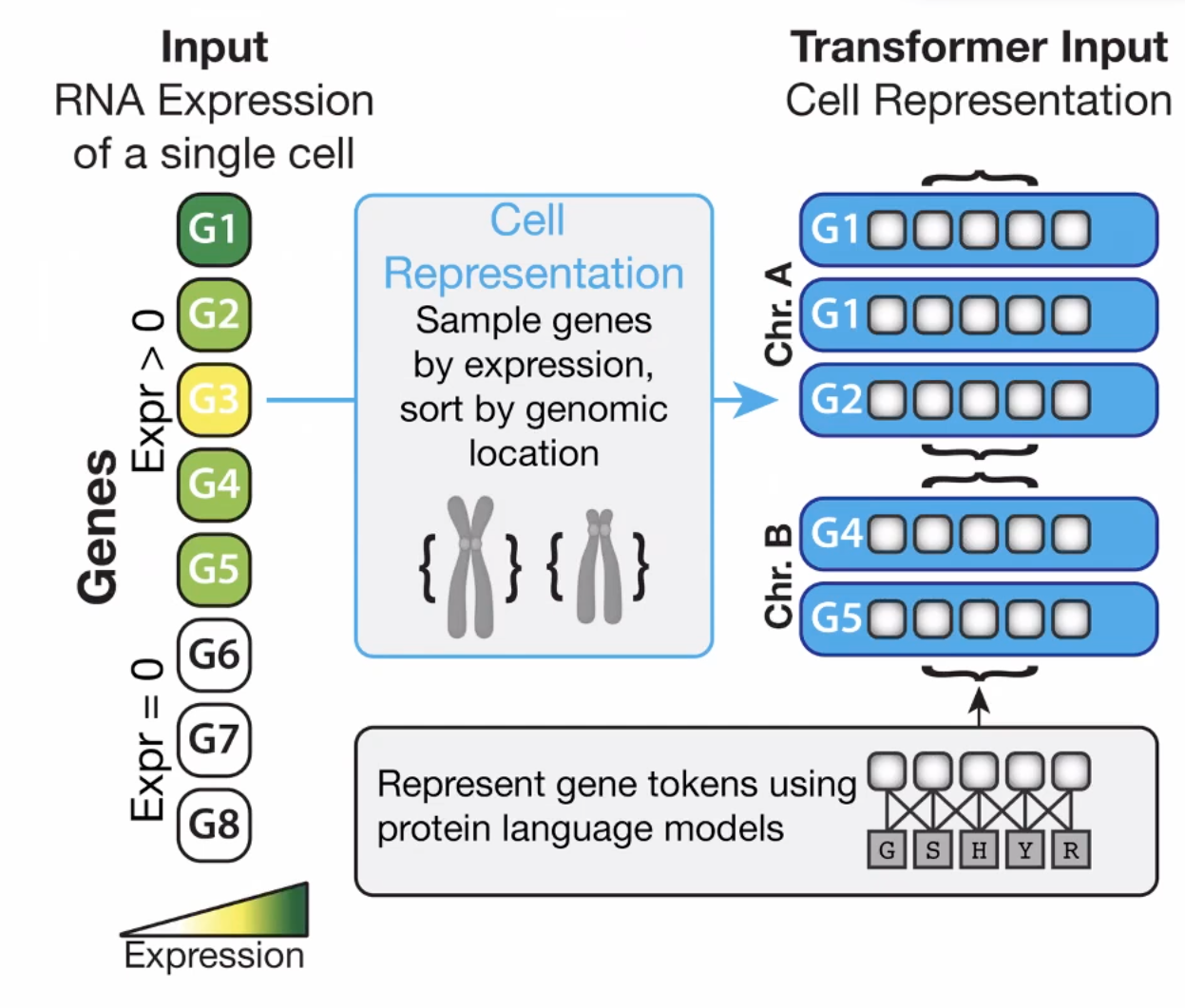 Output can be massive, and provides a universal space to be mapped
Output can be massive, and provides a universal space to be mapped
how high dimension is this bag of RNA representation space
TIME modality by ZMAN-seq from Ido Amit
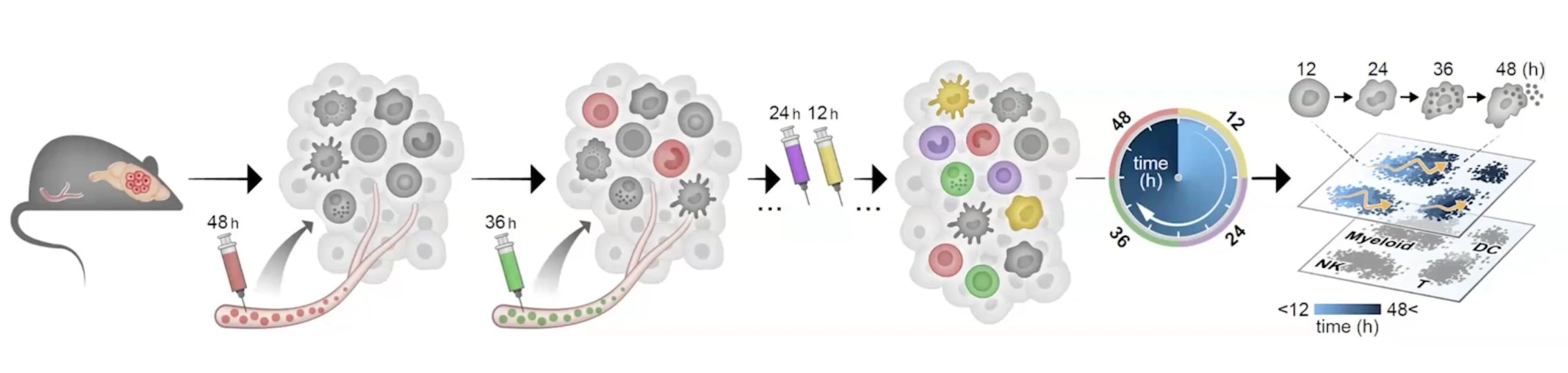
basically time stamped
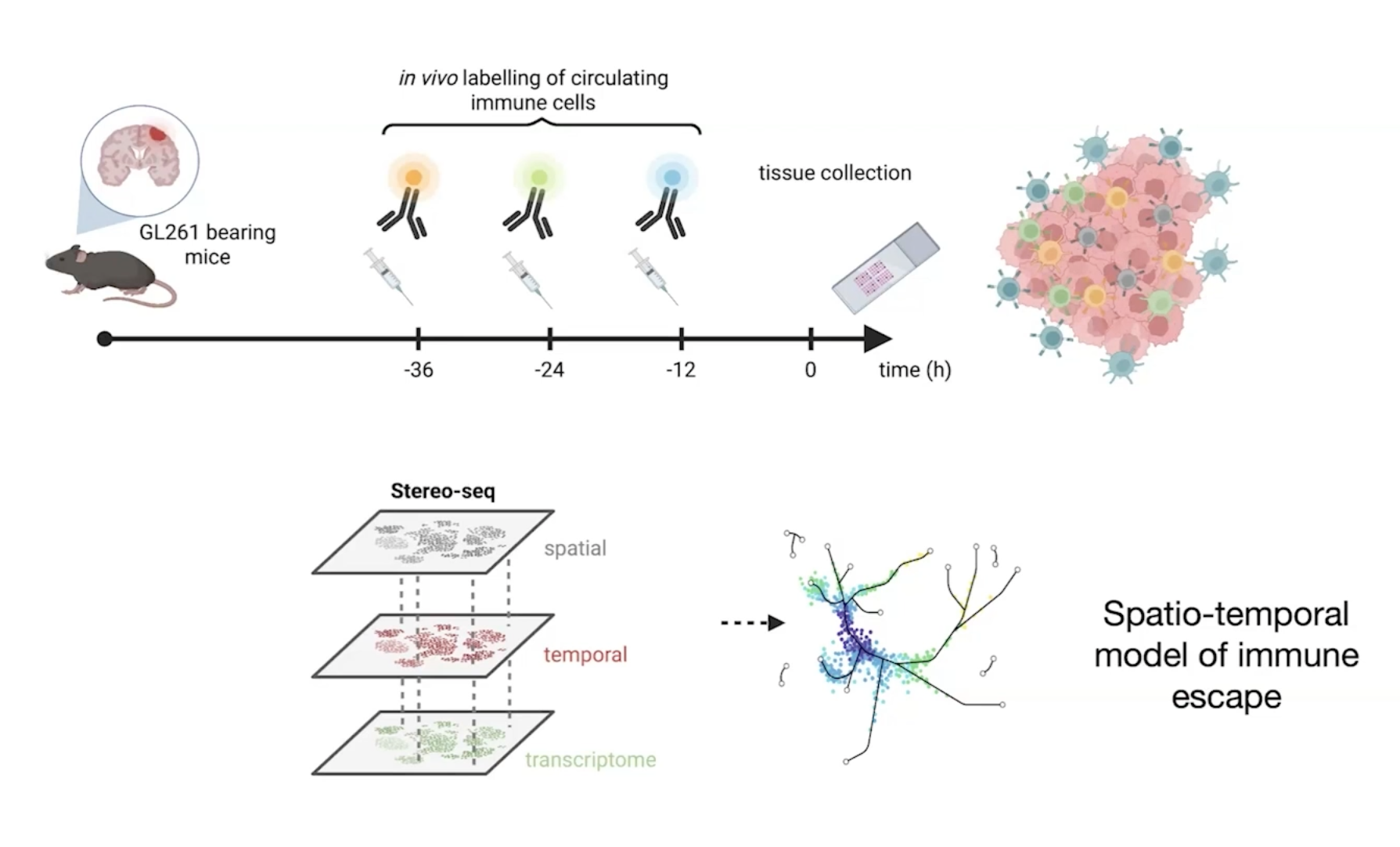
Makom & Zman-seq
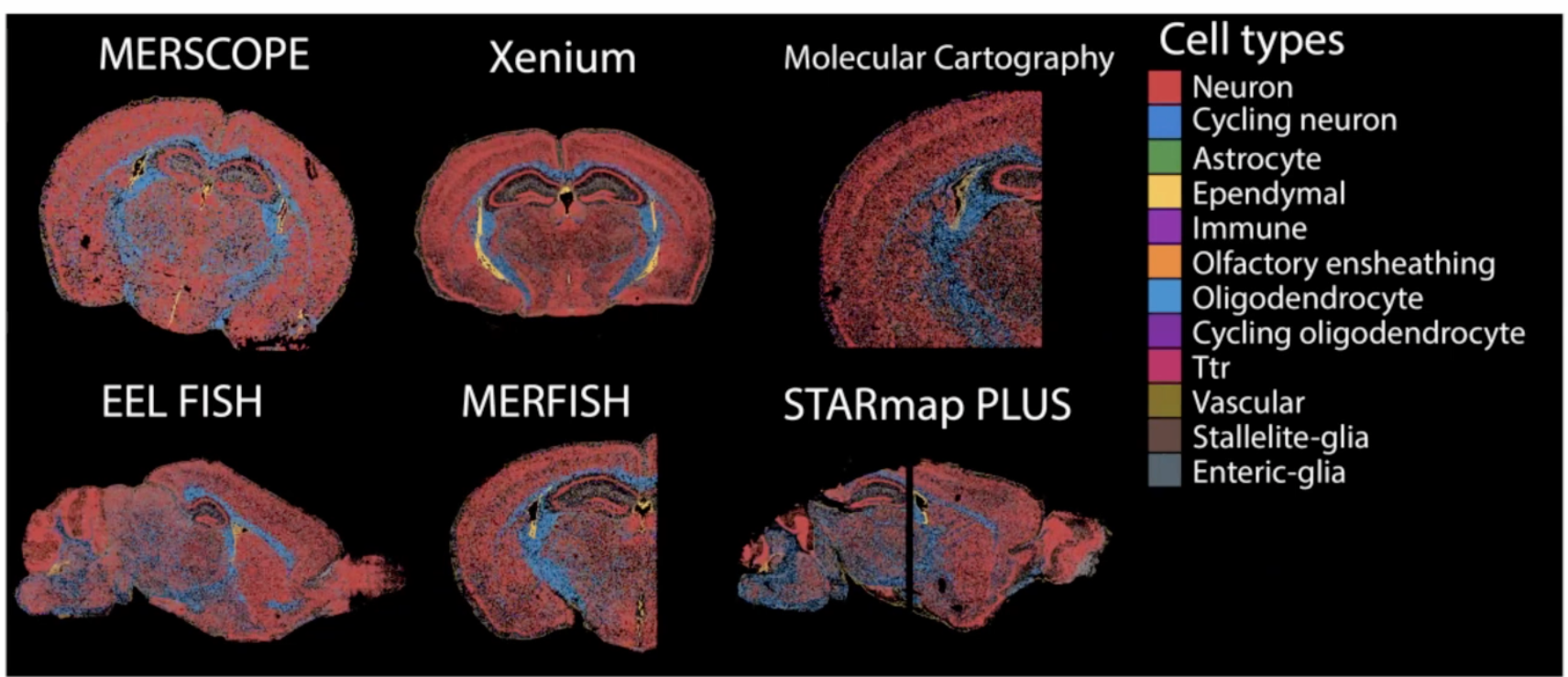
benchmark spatial seq techs
Methods of the Year 2020
Framework to identify Good data?
- targets not overlap across methods map to sc and benchmark sc
- spatial more abundant than sc, why
- lack of spcificity: barnyard-like plots to assess specificity
- lack of spcificity: coexpression of distinct cell type markers (mutually exclusive)
- confound with sensitivity: counts per cell correlate with segmentation size
- sensitivity and specificity bu controlling for segmentation differences
how?
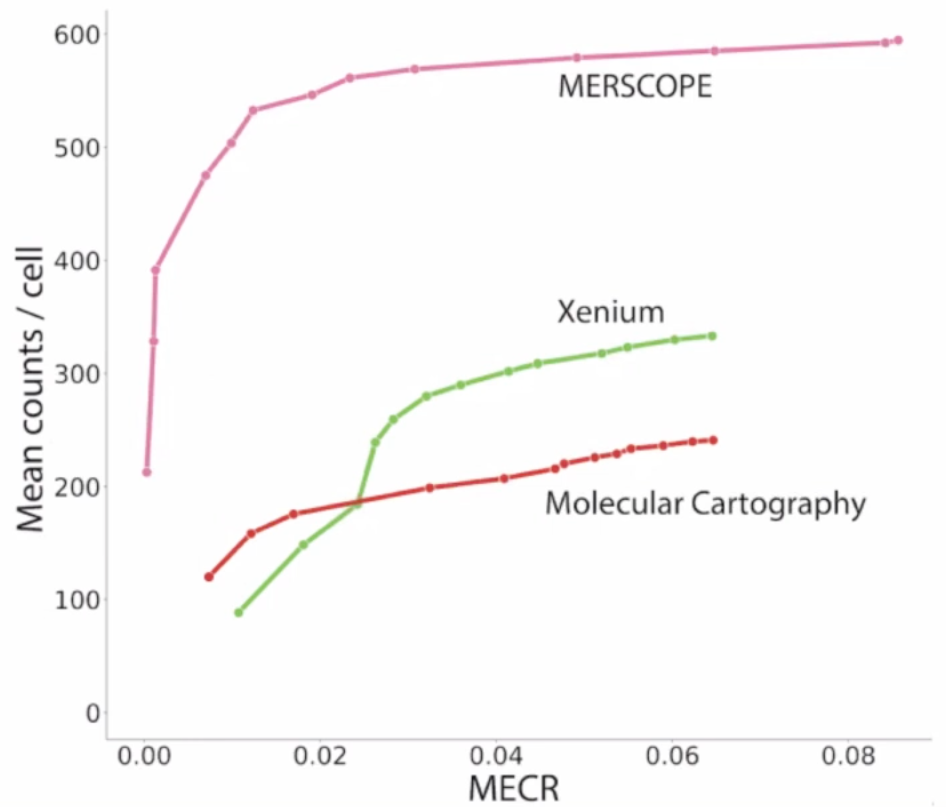
Qang, Huang, et al.,
Xenium, Merscope, CosMx from FFEP
- sensitivity: by spatial vs sc
- specificity: %genes above background [x] Xenium
Cook, et al.,
Spatial autocorrelation as a measure of quality. hypothesis: spatial patterning
- probe detection
- spatial patterning
- correlation with sc
- marker
open questions
- tardeoff panel size vs quality
- segmentation
- generality
if this is necessary?
Discussion
- segmentation problem, sts, molecules inaccuratelly assign is one reason, autofluorescence is another among many others
- recommend barnyard in mouse tissues (staining info from DAPI) is a good approach
Universal recording of cellular interactions by Sandra Nakandakari-Higa in Mucida Lab
LIPSTIC based on antigen on immune cells
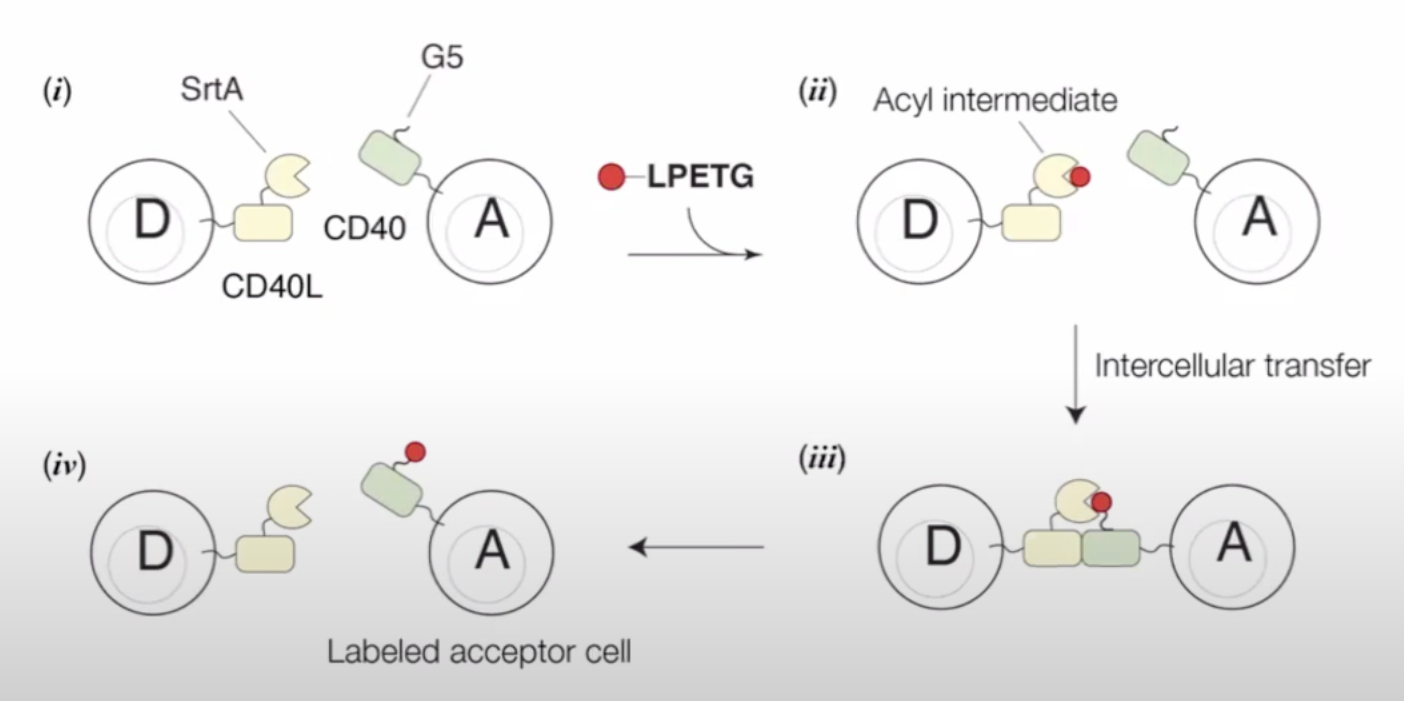
Universal LIPSTIC

this is good for study syncytiation
[x] quantitative measurement of interaction [x] outcome of interaction?
Disccusions
stability, label from 1-3 h depending on tissue and cell type
My impression
CHANLLENGES make this field worthy devoting